



Improving oedema disease diagnosis in pigs by detecting the vt2e toxin gen in oral fluid by qPCR
Here we report on the use of oral fluid (OF) and real time PCR (qPCR) as alternative methods in terms of sampling, representivness and cost.Introduction
Oedema disease caused by verotoxigenic strains of Escherichia coli (VTEC) is often diagnosed by bacterial culture from intestinal contents. This method is time consuming and requires additional tests to ensure that the isolate has the potential to produce the toxin. Here we report on the use of oral fluid (OF) and Real Time PCR (qPCR) as alternative methods in terms of sampling, representiveness and cost.
Materials and methods
A qPCR assay targeting the verocytotoxin variant 2e (vt2e) gene1 was standardised. A panel of 33 swine pathogens, and several 10-fold dilution series of a VTEC reference strain were used to determine the analytical performance of the assay. Then, the method was adapted to pig OF (image 1) as clinical sample to run two studies:
- Study 1: Detection of VTEC in OF. Diagnostic OF (n=99) came from nursery and fattening units in 18 commercial farms located in seven European countries. At each farm samples were collected from between two and twelve pens (x20 pigs per pen).
- Study 2: detection of VTEC-infected fattening unit. Samples were collected in a total of 16 pens as follows:
- swabs from floor, drinker and feeder (n=3 per pen);
- rectal swabs from all pigs present in each pen (n=12-16 per pen);
- OF (n=1 per pen).
Samples were processed in Diagnos (Hipra, Spain) by automatic DNA purification (Qiacube. Qiagen), and qPCR (Image 2). Results were compared in function of their origin (environment or animal).
Image 1. Oral Fluid sampling procedure: pigs chewing cotton rope
Image 2. Real time PCR instrument
Results
The qPCR detected the vt2e gene exclusively. The limit of detection was <1.5 ng of the target DNA/μg, and 9.6x103 CFU/ml. Efficiency was 93.3 percent.
Study 1:
vt2e was present in 17 of the 99 (17.2 percent) diagnostic OFs and 8 of the 18 (44.5 percent) farms (Figure 1).
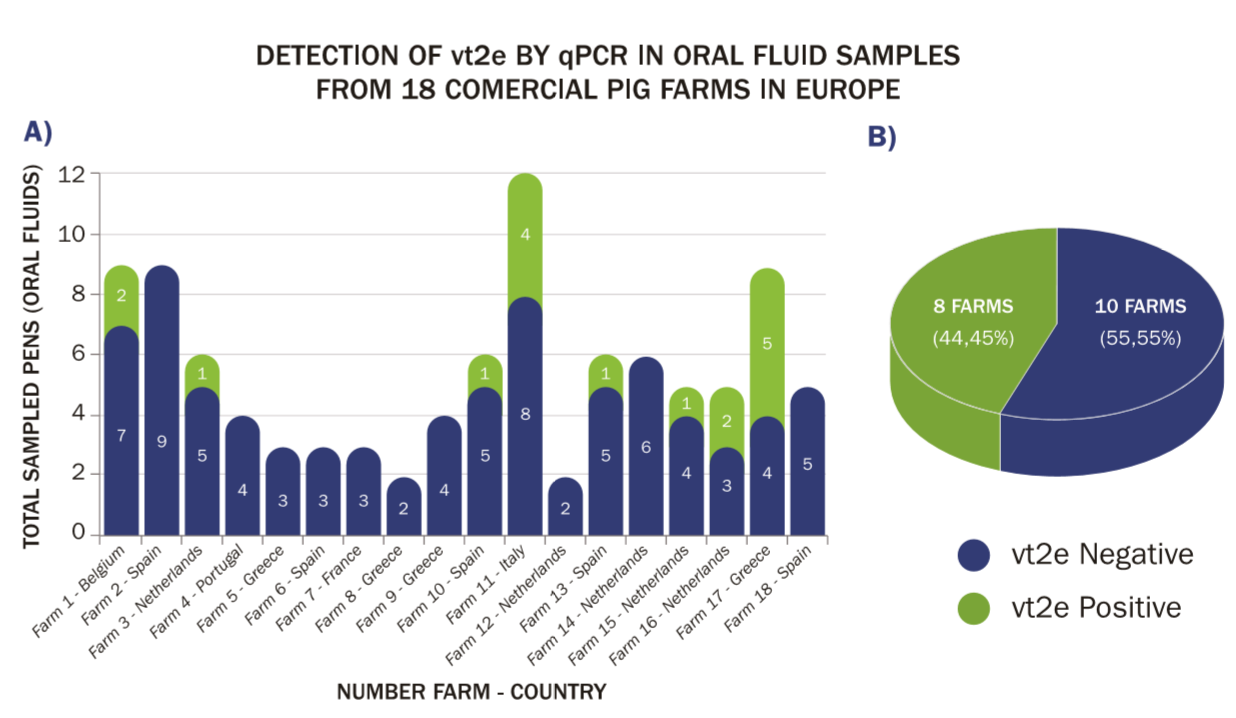
(A) Number and geographic origin of positive (green) and negative (blue) samples in 18 farms tested for the vy2e gene by qPCR.
(B) Percentage of vt2e positive and negative farms.
Study 2:
When analysing environmental, individual rectal swabs and collective OF samples coming from a VTEC-infected farm, 7/16 (43.5 percent), 12/16 (75 percent) and 16/16 (100 percent) were positive to vt2e, respectively (Table 1).
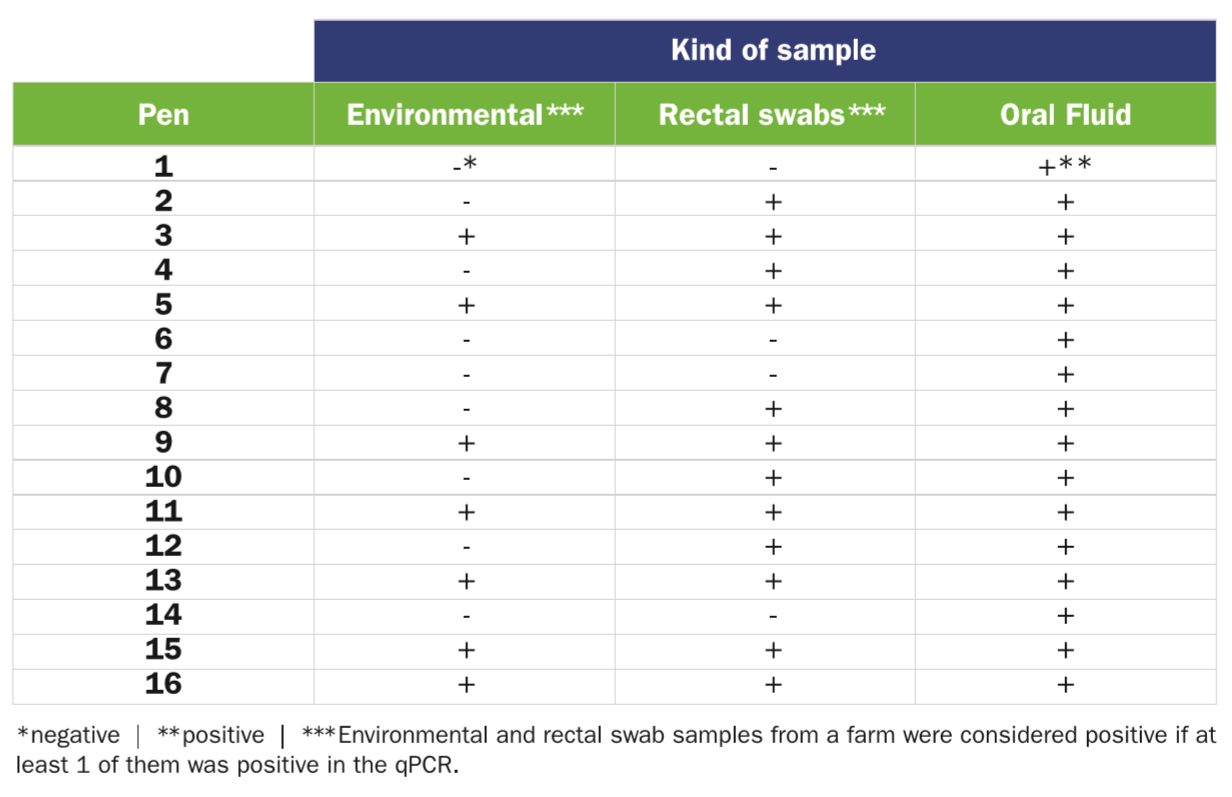
vt2e qPCR from 16 pens in a VTEC-infected fattening swine unit.
Discussion and conclusions
Results from this study demonstrate that the OF along with the qPCR can be useful alternative to bacteriology to monitor VTEC infection in pig herds. The proposed method reduces labour while increases the amount of animals sampled and reduces the waiting time to get results. It has been described that animals are more important reservoirs of VTEC than premises in the dairy farm environment2. Here we found similar results, indicating that hygiene and animal management are key in the control of oedema disease in pigs.
| References | ||||
|---|---|---|---|---|
| 1. Frydendahl, K et al. | ||||
| (2001) | . Molecular and cellular probes | 15, 151-160 | ||
| 2. Rahn, K et al. | ||||
| (1997) | . Epidemiol infect | 119:251-259 |







