



Pig Population Health Research Update
CANADA - Population health and the role genetics plays is a fascinating area of research and its implications for the industry can be substantial. New technology and computing power has allowed advancement in this area that previously was not possible and/or feasible, writes Nick Boddicker, Ph.D.The recent December issue of National Hog Farmer reported on key research carried out in 2015 and some of the articles included novel findings from large-scale PRRS virus (PRRSV) challenges.
Over the last 5 years there have been a number of large-scale disease challenge studies, including PRRSV and PCV2. These studies have identified animals that are less susceptible to these diseases through the use of genomics, transcriptomics (study of gene expression), and proteomics (study of protein expression).
Using this technology scientists have learned a great deal about the intricate relationships between pathogens and their hosts, specifically at the genetic level of the host (host genetics).
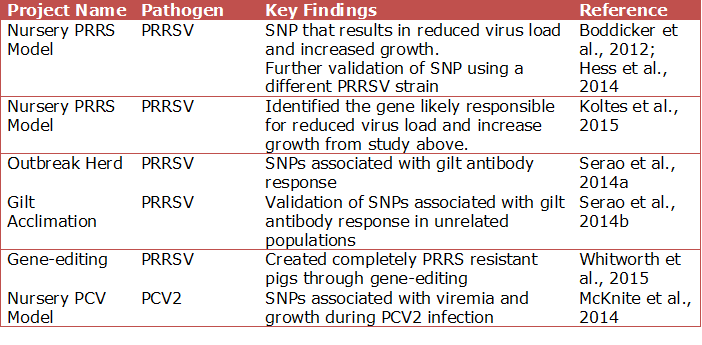
The projects summarized above have yielded promising results that could have a significant impact to the swine industry by reducing monetary losses during a PRRS or PCV2 outbreak, or even omitting them all together. Although quite novel, gene-edited animals have a long ways to go before they are put into production, if ever. Nonetheless, the gene-editing project revealed the mechanism of infection, could expedite methods to battle the virus more efficiently through genetics or pharmaceuticals, but will still take time.
Conversely, SNP information can be immediately incorporated into a selection program, but the effects of selection using the SNPs needs to be evaluated for affects on other economically important traits. For example, the SNP identified in the nursery PRRS model does not appear to be associated with growth or reproductive performance in non-challenged pigs in Genesus populations (Boddicker et al., 2014). Therefore, selecting for this SNP will not result in slower growing pigs or reduced reproductive performance when PRRS is not present, but can benefit the population during a PRRS outbreak. Similar results were found with one of the SNPs in the nursery PCV model.
However, when it comes to host genetics in response to a disease challenge there is a unique region on chromosome 7 that contains a cluster of immune response genes. This region is known as the Major Histocompatibility Complex (MHC). To effectively battle the array of swine diseases, this region innately contains a large amount of variation relative to the rest of the genome. In all the aforementioned projects, excluding gene-editing, important SNPs were identified within the MHC.
This is somewhat expected given the nature of this region, but it also begs the question “what do we do with it?”. By selecting on SNPs in the MHC, there is a chance of reducing the natural variation that exists, which could result in pigs that are less susceptible to e.g. PRRSV, but are not able to battle
influenza as effectively. This is a sophisticated region within the genome and further research is required to understand the genes and their effects associated with different pathogens.
With all the promising and exciting results from various disease challenges, it must come back to economics. The immune system is energy demanding and when activated results in reduced overall performance, a give and take relationship. Lochmiller and Deerenberg (2000) stated that the energetic costs of the immune system could be comparable to those of reproduction and growth.
Selecting for SNPs that are associated with the immune system could lead to naturally higher expression levels of immune-related genes resulting in higher maintenance requirements and reduced growth, feed intake and efficiency. Is it worth having less efficient animals in order to take less of a loss when an outbreak occurs? This is a question that still needs to be answered. Genesus continues to be involved in disease related projects and carries out additional research to investigate the effects of these SNPs on economically important production and reproduction traits before implementation into the selection program.
Finally, Genesus is collaborating in a new large-scale disease resilience study that is underway. This study entails the introduction of high-health pigs into a low-health grow-finish facility that contains an array of diseases found in the commercial setting. This study will attempt to identify SNPs that are associated with performance, feed efficiency and immune response in a low-health environment and not just response to a single pathogen. We will be reporting as results are available.
References:
Boddicker, N. J., E. H. Waide, R. R. R. Rowland, J. K. Lunney, D. J. Garrick, J. M. Reecy, and J. C. M. Dekkers. 2012. Evidence for a major QTL associated with host response to porcine reproductive and respiratory syndrome virus challenge. J. Anim. Sci. 90:1733–1746
Hess, AS, NJ Boddicker, RRR Rowland, JK Lunney, GS Plastow, JCM Dekkers (2014) A comparison of genetic parameters and effects for a major QTL between piglets infected with one of two isolates of porcine reproductive and respiratory syndrome virus. In: 2014 North American PRRS Symposium Proceedings, 68. North American PRRS Symposium, Chicago, IL, USA.
Koltes, JE, E Fritz-Water, CJ Eisley, I Choi, H Bao, A Kommadath, NVL Serao, NJ Boddicker, SM Abrams, M Schroyen, H Loyd, CK Tuggle, GS Plastow, L Guan, P Stothard, JK Lunney, P Liu, S Carpenter, RRR Rowland, JCM Dekkers, and JM Reecy. 2015. Identification of a putative quantitative trait nucleotide in guanylate binding protein 5 for host response to PRRS virus infction. BMC Genomics. 16:412.
Lochmiller, RL and C Deerenberg. 2000. Trade-offs in evolutionary immunology: just what is the cost of immunity. OIKOS. 88:87-98.
McKnite, AM, JW Bundy, TW Moural, JK Tart, TP Johnson, EE Jobman, SY Barnes, JK Qiu, DA Peterson, SP Harris, MF Rothschild, JA Galeota, RK Johnson, SD Kachman, and DC Ciobanu. 2014. Genomic analysis of differential response to experimental infection with porcine circovirus 2b. Animal Genetics. 45:205-214.
Serão, NVL, O Matika, RA Kemp, JCS Harding, SC Bishop, GS Plastow, JCM Dekkers. Genetic analysis of reproductive traits and antibody response in a PRRS outbreak herd J. Anim. Sci. 2014a. 92:2905–2921. doi:10.2527/jas.2014-7821
Serão, NVL, RA Kemp, B. Mote, JCS Harding, P. Willson, SC Bishop, GS Plastow, JCM Dekkers. 2014b. Whole-genome scan and validation of regions previously associated with PRRS antibody response and growth rate using gilts under health challenge in commercial settings. Proceedings 10th World Congress of Genetics Applied to Livestock production, Vancouver, BC.
Whitworth, K, RRR Rowland, CL Ewen, BR Trible, MA Kerrigan, AG Cino-Ozuna, MS Samuel, JE Lighner, DG McLaren, AJ Mileham, KD Wells, and RS Prather. 2015. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nature Biotechnology. Doi:10.1038/nbt.3434.







